Human Epidermal Organoids. Inflammatory Dermatoses. Cutaneous adverse drug reactions.
Skin Biology
The skin is our largest organ. Its barrier function is essential for the regulation of the body's water levels and temperature as well as for the protection from numerous environmental hazards including ultraviolet light, microbes, and chemicals. Under certain conditions, these agents can be pathogenic and lead to diseases, many of which remaining difficult to treat because of their recurrent or chronic nature as well as the lack of knowledge of their pathophysiology. Our research work aims to better understand pathologies targeting the epidermis with an emphasis on inflammatory conditions, i.e., cutaneous adverse drug reactions and rosacea, as well as alterations of the epidermis of genetic origin, such as Darier’s disease.
1. Study of severe drug reactions affecting the skin
In certain circumstances and individuals, drugs can trigger cutaneous inflammatory reactions whose severity can lead to the discontinuation of the culprit treatment. We have recently shown that certain drugs, such as amoxicillin, can induce the production of interleukin (IL)-36, a cytokine of the IL-1 family, resulting in the development of an acute cutaneous pustular reaction. We have also shown that IL-36 is involved in severe acne-like reactions in cancer patients treated with EGFR or MEK inhibitors.
Stevens-Johnson syndrome (SJS)/toxic epidermal necrolysis (TEN) is the most severe form of cutaneous adverse reactions and is induced, most of the time, by a medication. It is characterized by a skin detachment of variable extent due to a massive immune-mediated keratinocyte death. This keratinocyte death is the result of the cytotoxicity of drug-specific CD8+ T cells. To date, a highly efficient specific treatment to associate to the best supportive care is still badly needed as the mortality and the morbidity of SJS/TEN remains high. In collaboration with the European Reference Network-Skin (ERN-Skin), we aim to better understand the mechanisms involved in epidermal detachment with a special focus on innate immunity and IL-1 family cytokines. Recently, we have shown that IL-33, a cytokine/alarmin of the IL-1 family, can be found at high levels in the peripheral blood of SJS/TEN patients, which is most probably linked to a massive release by damaged epidermal keratinocytes. Interestingly, in the detaching epidermis in these patients, CD8+ T cells expressing the IL-33 receptor ST2L also express the cytolytic molecules granzyme B and perforin. This suggests that IL-33 may amplify the cytotoxic reaction involved in the keratinocyte death and the epidermis detachment. Our goal is to elucidate the impact of IL-33, and possibly other alarmins, on the keratinocyte-targeting immune response induced by a drug.
2. Study of chronic inflammatory diseases with the example of rosacea
Rosacea is an inflammatory skin disease affecting up to 60 million people worldwide. Due to the facial location of rosacea, many patients report stigmatization and psychological suffering. The current therapeutic options are aimed at dampening the inflammation, but a stable skin clearance is rarely achieved, and new drugs developed based on a better understanding of the pathophysiology are required. One element already known is that rosacea is driven by an exacerbated response mediated by LL-37, a factor normally released by cells from the epidermis to fight bacterial infections. However, how LL-37 impacts the immune response in the skin of patients suffering from rosacea remains incompletely understood. We recently found that certain intracellular signals are elevated in rosacea skin lesions. In particular, transcriptomic analyses of rosacea skin biopsies suggest that the JAK/STAT signaling pathway may be responsible for skin inflammation in rosacea. The activation of the JAK/STAT pathway may also be triggered by LL-37 in epidermal cells. Our work aims to identify whether the LL-37-JAK/STAT axis is a relevant inflammatory pathway in rosacea that could be addressed therapeutically. Indeed, several JAK/STAT inhibitors are under development by various companies and their potential in dermatology is undoubtedly very important.
3. Development of 3D epidermis models for the study of pathologies directly affecting the integrity of the epidermis
From the basal layer to the stratum corneum, the epidermis is composed of several layers of differentiating keratinocytes. Since keratinocytes exhibit various status in vivo, widely used monolayer keratinocyte cultures do not fully recapitulate the physiology of the epidermis. Darier’s disease is characterized by a defective keratinocyte adherence during their differentiation (acantholysis). To date, there is no safe and efficient treatment for this condition as well as no appropriate study model. With the objective of better understanding the molecular mechanisms involved in Darier's disease, we have first established Human Epidermal Organoid (HEO) cultures to recapitulate a healthy human epidermis in vitro. HEOs can be successfully generated from human foreskin keratinocytes, skin biopsies from healthy donors as well as from a keratinocyte cell line. HEOs resemble human epidermis as revealed by the expression of genes encoding collagen-4 and -17, keratin-1, -5, -10, -14 and -15, filaggrin, and transglutaminase-1 and -3. In addition, they retain low expression of the stem cell markers LGR5 and LGR6. We also demonstrated that these HEOs are reproducible on a large scale, allowing their use for high-throughput screenings. This model will be used to understand monogenic diseases manifesting by the loss of intercellular connections in the epidermis such as Darier’s disease.
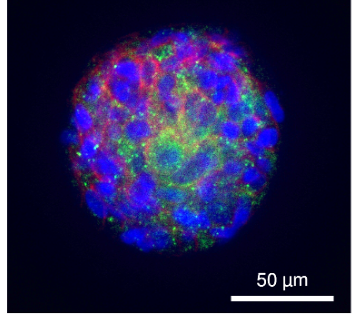
After 7 days of culture in Matrigel®, primary human keratinocytes form organized organoids. The illustration shows an HEO expressing the key desmosome proteins desmoglein-3 and desmoplakin.
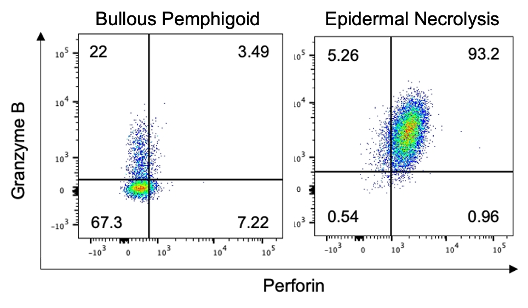
In epidermal necrolysis, blister CD8+ T cells expressing the IL-33 receptor ST2L also express the cytolytic molecules granzyme B and perforin. In contrast, CD8+ST2L+ cells from bullous pemphigoid, another blistering skin disease, do not exhibit a cytotoxic phenotype.
Social Media
