Neural Stem Cells . Neurogenesis . Notch Signaling . Post-transcriptional Gene Regulation Neurodegeneration . Brain Tumors
Embryology and Stem Cell Biology
Understanding brain development, maintenance and disease
Control of brain development from mice to human
How the billions of neurons and glial cells of the mammalian brain are generated from a pool of multipotent stem cells in a precise and orderly fashion is unclear. Using mouse genetics and human pluripotent stem cells (iPSCs) we are examining the molecular mechanisms controlling neural stem cell (NSC) maintenance, differentiation and fate choices. We perform proteomic and transcriptomic analysis of NSCs, progenitors and newly generated neurons at different stages of mouse development and from human iPSC cultures at the population and single cell levels and obtain unprecedented insight into the transcriptional dynamics and heterogeneity in stem and progenitor cells during neurogenesis.
Regulation of neurogenesis at the signaling and transcriptional levels
Neurogenesis continues in the adult mouse forebrain and hippocampus from NSCs, most of which are quiescent and only a few enter the cell cycle at any point in time. Notch signaling is critical for NSC maintenance. Of the four Notch proteins in mammals, we showed that Notch2 conveys quiescence to adult NSCs by regulating cell cycle-controlling genes and inducing expression of Id4, which prevents cell cycle entry. We found that Notch1 and Notch2 regulate different aspects of neurogenesis, therefore, we are comparing the regulome and interactome of Notch1 and Notch2 in the developing nervous system. In addition, we address the functions of other signaling pathways in the control of neurogenesis including Hippo signaling during cerebral cortex development. We analyze how different Tead transcription factors, the Hippo effectors, control different aspects of neurogenesis.
Regulating NSC fate at the post-transcriptional levels
The composition of a cell’s proteome determines its phenotype and function. Protein expression can be regulated at the mRNA level. We showed how the RNAseIII Drosha is an intrinsic regulator of adult NSC maintenance and differentiation though a novel mechanism of direct mRNA cleavage. Drosha represses expression of key transcription factors in NSCs by cleaving hairpins in their mRNAs. Intrigued by how a ubiquitous enzyme, Drosha, can cleave mRNAs in what we found to be a cell-type and stage-specific fashion, we use quantitative proteomics combined with co-immunoprecipitation and affinity pull-down assays to determine the Drosha interactome in NSCs. We identify novel protein complexes and examine how components of these complexes regulate Drosha function and cell fate.
Stem and progenitor cells and the origin of brain tumors
Gliomas are the most common primary brain tumors in adults and the prognosis in many cases is very poor. NSC-like cells exist in brain tumors leading to the question of whether they require the same molecular signals as normal NSCs. We are elucidating molecular mechanisms underlying transformation of neural stem and progenitor cells into tumor cells. Notch signaling has often been implicated as an oncogenic pathway in glioma. Contrary to predictions, we have uncovered a tumor suppressive function for the Notch pathway in brain tumor subtypes. We are currently addressing the differences and similarities between glioma stem cells and NSCs on the molecular level.
Modelling human disease with iPSCs
Human iPSCs can be generated from normal or diseased individuals and retain the ability to generate all cell-types of the adult organism. We are developing differentiation protocols to drive iPSCs into specific neural lineages. We are able to isolate and purify brain region-specific progenitors, neurons and astrocytes from iPSC-derived cultures over >100 days of differentiation. Recently, we established highdensity micro-electrode arrays to monitor neuronal circuit formation and monitor disease phenotypes in human iPSC-derived neurons. We have generated iPSC from patients suffering from neurodevelopmental and neurodegenerative disorders including Parkinson’s disease, FTLD, and MYCOPS12. Using iPSCs as a model system and the directed differentiation to neurons and glia, we are studying the effects of genetic mutations on neuronal cell biology, gene expression and neuronal function.
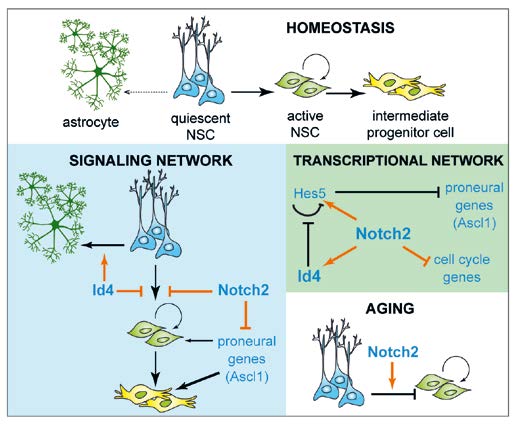
Fig. 1: NSCs of the adult hippocampus are multipotent but mitotically inactive. They must enter cell cycle to generate neurons during homeostatic maintenance and they transit to astrocyte production with age. Notch2 blocks NSC entry into the cell cycle by inducing Id4 expression, and inhibits Ascl1 transcription, thereby simultaneously preventing neuronal fate commitment. Id4 increases Hes5 expression by blocking its autorepression, and promotes astrocyte differentiation of NSCs. Increased Notch2 activity in ageing NSCs contributes to reduced neuron production and commitment to astrocytes differentiation.
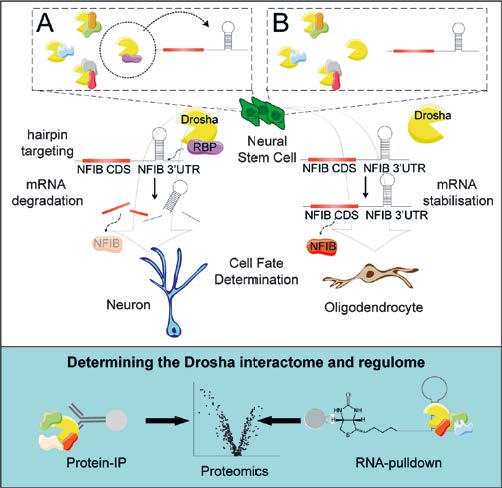
Fig. 2: Drosha regulates NSC fate by targeted cleavage of mRNAs of fate determining transcription factors. NFIB drives oligodendrocyte commitment of hippocampal NSCs, but its expression is repressed but Drosha cleavage of its mRNA. A. Cell-type specific regulation of Drosha activity is controlled by expression of cell-specific RNA binding protein (RBP) partners which target mRNAs in a sequence-specific fashion. B. Down-regulation of Drosha partners stabilizes the mRNA of targeted fate determining factors. Quantitative proteomics identified more than 140 novel Drosha interacting proteins and proteins that bind Drosha mRNA targets.
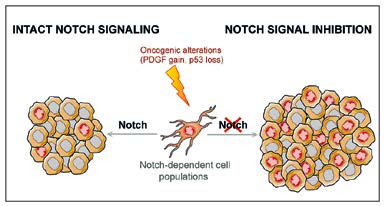
Fig. 3: Gliomas are the most common primary brain tumors in adults and the prognosis for patients is, in many cases, very poor. It has been proposed that gliomas could aberrantly hijack the same signaling pathways that allow normal NSCs to self-renew, thus enabling glioma cells to proliferate extensively and form the tumor. Unexpectedly, our studies indicate that Notch signaling, one major molecular pathway implicated in NSC self-renewal, can actually suppress the formation of some forms of brain tumor.
Social Media
