Detection and Outcome Markers . Metastasis . EMT . Aberrant Glycosylation, Drugs and Treatment . Therapeutic Targets
Ovarian Cancer Research
Ovarian cancer (OC) is the most lethal gynecological malignancy and the 5th leading cause of cancer death in women. Owing the lack of reliable diagnostic markers, OC is typically diagnosed at late stage, when it has already spread into the peritoneal cavity and complete surgical removal is difficult.
The etiology of OC is not fully understood despite several decades of intensive research. OC may arise from transformed epithelial cells in the fallopian tube rather than from the ovarian surface epithelium. At risk are women with germ line mutations in BRCA genes and somatic mutations in BRCA andTP53.
Clinical management of patients with OC is by cytoreductive surgery and chemotherapy and rarely radiotherapy; immune therapeutic approaches are currently under clinical evaluation. But therapy failures and recurrence are often observed. A solution to this problem is offered by personalized (precision) medicine, where the therapy is precisely tailored to the molecular signature of the patient’s tumor.
Metastasis is apart from therapy failure and resistance development a major obstacle in OC treatment. The preferred site of OC metastasis is the peritoneal cavity and in particular the omentum, a layer of fatty tissue that also contains various cell types including immune cells. Metastasis can occur via different routes (lymphatic spread, hematogenous spread, and passive dissemination via ascites flow) and involves the shedding of tumor cells from the primary tumor and the establishment of the secondary tumor (metastasis) at distant sites via orchestrated but not poorly understood processes.
The Ovarian Cancer Research (OCR) Group, run by groupleader Prof. Dr. med. Viola Heinzelmann-Schwarz and co-groupleader PD Dr. sc. nat. Francis Jacob, is a highly motivated team of complementary expertise dedicated to the better understanding of the molecular nature of this disease and to contribute to the well-being of the patients suffering from it.
Research Focus
- Identification of predictive biomarkers and molecular signatures (genetic and non-genetic) for early detection of ovarian cancer and treatment response prediction
- Function of protein and lipid glycosylation in ovarian cancer development, progression, metastasis, and therapy response
- Establishment of experimental assays and ex vivo patient-derived xenograft models for (personalized) treatment prediction
- Identification of molecular strategies to tackle/circumvent drug resistance in ovarian cancer
Current Research
Establishment of an assay to predict treatment response in ovarian cancer based on a novel functional homologous recombination (HR) assay: Treatment regimens in oncology frequently become personalized and multi-modal, but reliable biomarkers or measures that allow the accurate prediction of therapy responses in individual patients are scarcely available. In this project we intend to establish and validate a highly reliable in vitro assay that predicts treatment response (radio- and chemotherapy) in ovarian cancer cell lines as well as in cultures of patient-derived tumor cells.
Establishment of in-house established CRISPR-Cas9 cell competition assay as a novel tool to identify predictive biomarkers for PARPi response: To be able to identify OC patients that will truly benefit from PARPi and also to better understand the molecular mechanisms underlying the response/non-response, we have established and currently validate an experimental in vitro CRISPR-Cas9 mutagenesis assay to functional validate candidate genes in PARPi sensitivity. This assay not only validates genes for PARPi response (and perhaps to other classes of DNA-damaging agents) but also allows the functional assessment of the role of specific genes in cancer cell proliferation/survival identifying possible vulnerabilities for cell survival and therefore potential candidates for targeted therapy.
Zebrafish embryo tumor xenograft and ex vivo 3D culture models for functional drug testing, patient specific predictions, and personalized therapies in ovarian cancer: We aim at developing, establishing, and validating a Zebrafish (patient-derived) tumor xenograft (zPDX) model that can be used to study cancer metastasis (replacing the mouse model) and as an avatar to predict therapy responses in ovarian cancer patients. Obtained data are then compared with the therapy response and related to the clinic pathological data of the patients. Moreover, we aim at designing the experimental protocol and setup to establish a scaffold-free 3D culture model for (as a first approach) ovarian cancer cell lines and subsequently for patient-derived tumor cells (proof-of-protocol). This includes the generation of 3D cell aggregates, FFPE-embedding, and immunofluorescence staining with the markers specific for the various cell types (cancer cells, immune cells, fibroblasts) and cell states (proliferative, apoptotic) imaging and, determination of the relative composition of the cell types/states according to the relative abundance of the markers (readout). This ex vivo model is expected to perform with high efficacy regarding its capacity to generate ex vivo cultures from ovarian cancer patient-derived tumor cells and hence to capture the expression profiles of matched tumor samples and exhibit heterogeneous interpatient drug responses.
SMAC mimetics: an approach to overcome ovarian cancer chemo-resistance (by targeting the MSLN-TMEM100-TNFalpha axis): In this project, we hypothesize that SMAC mimetics, chemical antagonists of inhibitor of apoptosis proteins (IAPs), are specifically effective in targeting chemo-resistant ovarian cancer cells. Our preliminary data indicate that the combination of first-line chemotherapeutic compounds with the SMAC mimetics is synergistically cytotoxic in ovarian cancer cell line models. We are currently exploring the molecular nature behind this synergism and the underlying pathways, and will as a next step try to recapitulate this finding in an in-house developed ex vivo patient derived tumor model and in a nude mice model.
Single-cell multimodal expression profiling technology as an approach to understanding the role of the tumor microenvironment and its changes during ovarian cancer progression and recurrence: We use single-cell multimodal expression profiling technology to longitudinally profile the molecular changes in the ascites, from initial diagnosis to recurrence. Ascites is a protein- and metabolite-rich abdominal fluid “ecosystem” often found in ovarian cancer patients. This technology integrates the transcriptome and the surfaceome (glycoproteins and glycan proteome, and glycome on a single-cell level, enabling to monitoring of non-genetic changes that may accompany disease progression, recurrence, and therapy resistance.
The Swiss Tumor Profiler: multi-modal cancer profiling in patients with high-grade serous ovarian cancer: The multi-centric (among others: University Hospitals of Basel and Zurich, ETH, University of Zurich) Swiss Tumor Profiler (TuPro) is an innovative, interdisciplinary, and multi-technology-based observational trial that integrates in-depth genetic and molecular tumor profiling. The aim of TuPro is also to better identify and predict successful treatment targets in order to develop new molecular-based therapies. This multi-modal profiling includes a variety of platforms, many of them are based on single-cell level analyses. The computational integration of these data allows to support clinicians in making patient-specific treatment decisions and thus creates novel opportunities for personalized medicine. Clinical, logistical, experimental, and computational workflows to achieve the analyses of small cancer biopsies or ascites/blood in a clinical setting are meanwhile established (Irmisch et al, 2021; Stark et al, 2021).
Clinical Research and Clinical Trials: An overview of further ongoing clinical projects/trials can be found on the website of the CTU (Departement Klinische Forschung) of the University Hospital Basel
and on the website of the Swiss GO Trial Group
https://swiss-go.ch/de/klinische-studien
Terminated Research
PARP-inhibitor (PARPi) resistance and experimental approaches towards its evasion in ovarian cancer cells: Intrinsic and acquired resistance chemotherapeutics including PARPi is a major obstacle in cancer treatment. We have recently shown that PARPi are unlikely inducers of PARPi-resistance (Fedier et al, 2022). Here, we aim identifying using our in-house developed CRISPR-Cas9 cell competition assay genetic markers, i.e. mutations in genes other than BRCA1/2, that can re-sensitize chemo-resistant ovarian cancer cells to PARPi. The identified genes, i.e. in their mutated and loss-of-function variant, are potential candidate markers to predict intrinsic and acquired PARPi response in tumor cells. This project may also uncover so far unknown genes linked to homologous recombination (HR) repair-dependent cellular responses to DNA damage. We were able to show that that besides the obvious candidates BRCA1 and BRCA2, also ATM, MUS81, NBN, RAD51, and other genes were identified as potential predictor genes for PARPi response (Coelho et al, 2022).
Ovarian cancer detection and outcome markers: MELK (embryonic leucine-zipper kinase) expression was identified to correlate with poor survival in ovarian cancer (Kohler et al, 2017), whereas expression of LATs was not associated with outcome in ovarian cancer patients (Montavon et al, 2019). A preceding study characterized the N- and O-glycome of ovarian cancer (OC) and peritoneal cancer (PC) using tissue glycomics and revealed distinct glycomic signatures, i.e. proteins are differently and uniquely glycosylated in these two cancers (Anugraham et al, 2017). A more recent study using transcriptomic and next-generation sequencing data and validation by immunohistochemistry showed that OC and PC are epidemiologically and molecularly distinct diseases: PC relapsed earlier, had a distinctively different gene signature, and showed a different sensitivity to standard chemotherapy drugs compared to OC (Jacob et al, 2020).
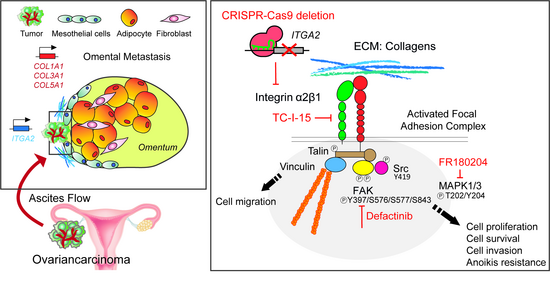
Establishment of a 3D human omentum model to study ovarian cancer metastasis and its molecular regulators: Loss of globoside glycosphingolipids by genomic deletion of A4GALT induced an epithelial-to-mesenchymal transition (EMT), in OC cells, associated with loss of E-cadherin expression and cell-cell adhesion and with increased chemoresistance (Jacob et al, 2018). EMT is thus functionally linked and regulated by dynamic alterations in lipid glycosylation (Cumin et al, 2021, 2022). In a collaborative project, we have identified and molecularly characterized the cell types composing human omentum and established a functional 3D-bioprinted omentum model to study the mechanisms of OC metastasis to the omentum (peritoneum) (Estermann et al, 2023). This project (Huang et al. Elife. 2020) highlights the significance of the omentum in OC metastasis: when enriched in collagen, it presents a pre-metastatic niche for tumor cells to further develop into peritoneal metastases. It also highlights the importance of integrinA2 (ITGA2) in metastasis and proposes a new route of metastasis through the interaction of ITGA2 with collagens. In addition, we found that mesothelin (MSLN) promotes invasion of tumor cells through the mesothelial cell layer in vitro, suggesting MSLN as key player in OC progression by triggering peritoneal dissemination (Coelho et al, 2020).
Selected Publications (last 5 years)
See separate listing. https://pubmed.ncbi.nlm.nih.gov/?term=Heinzelmann-Schwarz+V&sort=date&size=20
Selected Collaborations/Network
- Institute Adolphe Merkle, University of Fribourg, (Prof. Barbara Rothen-Rutishauser). 3D bioprinting models.
- FHNW University of Applied Sciences and Arts Northwestern Switzerand, Muttenz, (Prof. Eric Kübler, Prof. Laura Suter-Dick). DNA and RNA diagnostics; cell culture and tissue engineering.
- ETH Basel (Dr. Christian Beisel and Dr. Franziska Singer (NEXUS). Single Cell Technologies.
- Paul Scherrer Institute PSI, Villingen (Prof. Jürgen Grünberg, Dr. Martin Behe). Ovarian cancer metastasis in mouse models and design of radionuclid- and drug-coupled antibodies for therapy.
- Tumor Profiler (TuPro) Consortium (University Basel, University Zurich, ETH Zurich, Swiss Institute of Bioinformatics, Roche Innovation Center Zurich). An interdisciplinary and interinstitutional project aimed at the molecular dissection of melanoma, ovarian cancer, and AML in order to genetically and molecularly dissect the tumors on single-cell level and to support clinicians in the best choice of therapy.
- University of Porto, Portugal (Prof. Leonor David). Mesothelin in cancer progression and metastasis.
- University of Copenhagen, Denmark (Dr. Henrik Clausen). Biology of E-cadherin glycosylation.
- University Medical Center Hamburg-Eppendorf, Germany (Prof. Tobias Lange). Integrin glycosylation in mouse metastatic xenograft models.
- Leibniz Research Centre for Working Environment and Human Factors, Dortmund, Germany (Dr. Jan Hengstler). Molecular mechanisms of ascites development.
- Griffith University, Gold Coast, Australia (Prof. Daniel Kolarich, Dr. Arun Everest-Dass, Prof. Mark von Itzstein). Analysis of glycosylated key cell surface proteins in metastasis.
- Quantgene Inc., Delaware Corporation, Berkeley, CA, USA. A feasibility study for investigating circulating tumor DNA (ctDNA) exposure in peripheral blood using a novel process.
Specilities and Ressources
We have a large biobank (BOB, Basel Ovarian Biobank) with blood, tissue (frozen and embedded, tissue microarrays), ascites, urine, and primary tumor cells (frozen) from over 3’500 ovarian cancer patients from 3 different locations (Sydney AUS, Zurich and Basel), including matched and longitudinal samples. The biobank also includes breast milk samples (frozen) from a Swiss and a Mongolian cohort.
We have methodological expertise in molecular and translational cancer biology, glycobiology, and drug resistance; and employs modern lab technologies including CRISPR-Cas9-genome editing (silencing, activation, knock-out, knock-in, and library screens), fluorescence and live cell imaging, cultivation of primary cultures, zebrafish embryo tumor xenografts, ex vivo xenograft models, multi-color fluorescence activated cell sorting. We tightly collaborate with experts in single cell analyses and bioinformatics.
Current Grants and Support
- Swiss National Science Foundation (Molecular profiling and drug prediction in ovarian cancer patients). 2022-2026.
- Krebsforschung Schweiz (Innovative ovarian cancer zPDX models for prediction of treatment regimen outcomes in precision medicine). 2022-2025.
- Tumor Profiler Study (Interdisciplinary and interinstitutional project aimed at the molecular dissection of melanoma, ovarian cancer, and AML in order to genetically and molecularly dissect the tumors on single-cell level and to support clinicians in the best choice of therapy; supported a.o. by the Universities of Basel and Zurich, ETH Zurich, Roche Innovation Center Zurich). 2019-2024.
- PHRT ETH Pioneer Project Grant (Dissect longitudinal evolutionary trajectories in ovarian cancer patients using integrated proteogenomics). 2020-2022.
- MATAO trial; MAintenance Therapy with Aromatase inhibitor in epithelial Ovarian cancer: a randomized double-blinded placebo-controlled phase III trial (support: a.o. Swiss GO Trial Group, Helsana Krankenkasse; Krebsforschung Schweiz, Anticancer Fund/Reliable Cancer Therapies, Belgium/Switzerland; Stiftung Fürstlicher Kommerzienrat, Liechtenstein). 2019-2024.
- Wilhelm Sander-Stiftung (Influence of glycosphingolipids on molecular and cellular mechanisms in metastatic ovarian cancer). 2019-2022.
Social Media